HomeWork #1 Machine Learning
Contents
HomeWork #1 Machine Learning#
Stu. name: Seyed Mohammad Amin Dadgar
Stu. no: 4003624016
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
import warnings
import scipy
Q2#
## column names are used from ```pima-indians-diabetes.name``` file
cols = ['pregnancy_count', 'glucose_test', 'blood_pressure', 'triceps_thickness', '2h_insulin', 'mass', 'pedi', 'age', 'label']
df = pd.read_csv('hw1_data/pima/pima-indians-diabetes.data', index_col=False ,names=cols)
df.head()
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
## save the dataset in the right format with its columns for other usages
df.to_csv('hw1_data/processed/pima-indians-diabetes.csv')
(a, b)#
df.describe()
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| count | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 |
| mean | 3.845052 | 120.894531 | 69.105469 | 20.536458 | 79.799479 | 31.992578 | 0.471876 | 33.240885 | 0.348958 |
| std | 3.369578 | 31.972618 | 19.355807 | 15.952218 | 115.244002 | 7.884160 | 0.331329 | 11.760232 | 0.476951 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.078000 | 21.000000 | 0.000000 |
| 25% | 1.000000 | 99.000000 | 62.000000 | 0.000000 | 0.000000 | 27.300000 | 0.243750 | 24.000000 | 0.000000 |
| 50% | 3.000000 | 117.000000 | 72.000000 | 23.000000 | 30.500000 | 32.000000 | 0.372500 | 29.000000 | 0.000000 |
| 75% | 6.000000 | 140.250000 | 80.000000 | 32.000000 | 127.250000 | 36.600000 | 0.626250 | 41.000000 | 1.000000 |
| max | 17.000000 | 199.000000 | 122.000000 | 99.000000 | 846.000000 | 67.100000 | 2.420000 | 81.000000 | 1.000000 |
## the variance of each attribute
pd.DataFrame(df.var(), columns=['variance'])
| variance | |
|---|---|
| pregnancy_count | 11.354056 |
| glucose_test | 1022.248314 |
| blood_pressure | 374.647271 |
| triceps_thickness | 254.473245 |
| 2h_insulin | 13281.180078 |
| mass | 62.159984 |
| pedi | 0.109779 |
| age | 138.303046 |
| label | 0.227483 |
(c)#
## Calculating the correlation between 8 attributes (label is outcluded)
attr_corr = df[cols[:-1]].corr()
attr_corr
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | |
|---|---|---|---|---|---|---|---|---|
| pregnancy_count | 1.000000 | 0.129459 | 0.141282 | -0.081672 | -0.073535 | 0.017683 | -0.033523 | 0.544341 |
| glucose_test | 0.129459 | 1.000000 | 0.152590 | 0.057328 | 0.331357 | 0.221071 | 0.137337 | 0.263514 |
| blood_pressure | 0.141282 | 0.152590 | 1.000000 | 0.207371 | 0.088933 | 0.281805 | 0.041265 | 0.239528 |
| triceps_thickness | -0.081672 | 0.057328 | 0.207371 | 1.000000 | 0.436783 | 0.392573 | 0.183928 | -0.113970 |
| 2h_insulin | -0.073535 | 0.331357 | 0.088933 | 0.436783 | 1.000000 | 0.197859 | 0.185071 | -0.042163 |
| mass | 0.017683 | 0.221071 | 0.281805 | 0.392573 | 0.197859 | 1.000000 | 0.140647 | 0.036242 |
| pedi | -0.033523 | 0.137337 | 0.041265 | 0.183928 | 0.185071 | 0.140647 | 1.000000 | 0.033561 |
| age | 0.544341 | 0.263514 | 0.239528 | -0.113970 | -0.042163 | 0.036242 | 0.033561 | 1.000000 |
## showing the heatmap for better visualization
sns.heatmap(attr_corr)
plt.show()
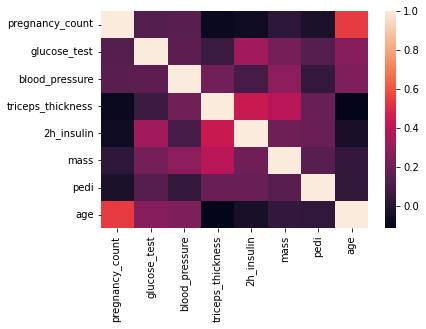
label_corr = df.corrwith(df.label).sort_values(ascending=False)
pd.DataFrame(label_corr, columns=['correlation'])
| correlation | |
|---|---|
| label | 1.000000 |
| glucose_test | 0.466581 |
| mass | 0.292695 |
| age | 0.238356 |
| pregnancy_count | 0.221898 |
| pedi | 0.173844 |
| 2h_insulin | 0.130548 |
| triceps_thickness | 0.074752 |
| blood_pressure | 0.065068 |
What we will find out from the correlation matrix with label is the most useful feature (Or the most affective feature ) for the label is glucose_test. To find why it is the most helpful attribute we can review the correlation formula with below \begin{equation} correlation = \frac{covariance(x,y)}{var(x) var(y)} \end{equation} And with this equation it’s obvious that if coefficient is positive then the effect of can be if x increases then y is increased. By this reason the biggest value for correlation of a feature with lable can be intrepreted the most helpful feature (attribute).
Also to explain the correlation value of one we can say that, x variable is the same as y in eq(1).
(d)#
If 2 attributes are fully correlated then using both in prediction may make bias and the prediction result would not be helpful. The alternative and the better way for prediction is to use one of the attributes.
(f)#
fig, ax = plt.subplots(3, 3, figsize=(15,9))
fig.tight_layout(pad=5)
sns.distplot(df.glucose_test, bins=20, ax=ax[0,0])
ax[0,0].set_title('Plasma glucose concentration a 2 hours\n in an oral glucose tolerance test')
sns.distplot(df.mass, bins=20, ax=ax[0,1])
ax[0, 1].set_title('Body mass index \n(weight in kg/(height in m)^2)')
sns.distplot(df.age, bins=20, ax=ax[0,2])
ax[0, 2].set_title('Age')
sns.distplot(df.pregnancy_count, bins=20, ax=ax[1,0])
ax[1, 0].set_title('Number of times pregnant')
sns.distplot(df.pedi, bins=20, ax=ax[1, 1])
ax[1, 1].set_title('Diabetes pedigree function')
sns.distplot(df['2h_insulin'], bins=20, ax=ax[1, 2])
ax[1, 2].set_title('2-Hour serum insulin (mu U/ml)')
sns.distplot(df.triceps_thickness, bins=20, ax=ax[2, 0])
ax[2, 0].set_title('Triceps skin fold thickness (mm)')
sns.distplot(df.blood_pressure, bins=20, ax=ax[2, 1])
ax[2, 1].set_title('Diastolic blood pressure (mm Hg)')
sns.distplot(df.label, bins=20, ax=ax[2, 2])
ax[2, 1].set_title('Class variable (0 or 1)')
plt.show()
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
/home/amin/.local/lib/python3.8/site-packages/seaborn/distributions.py:2619: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
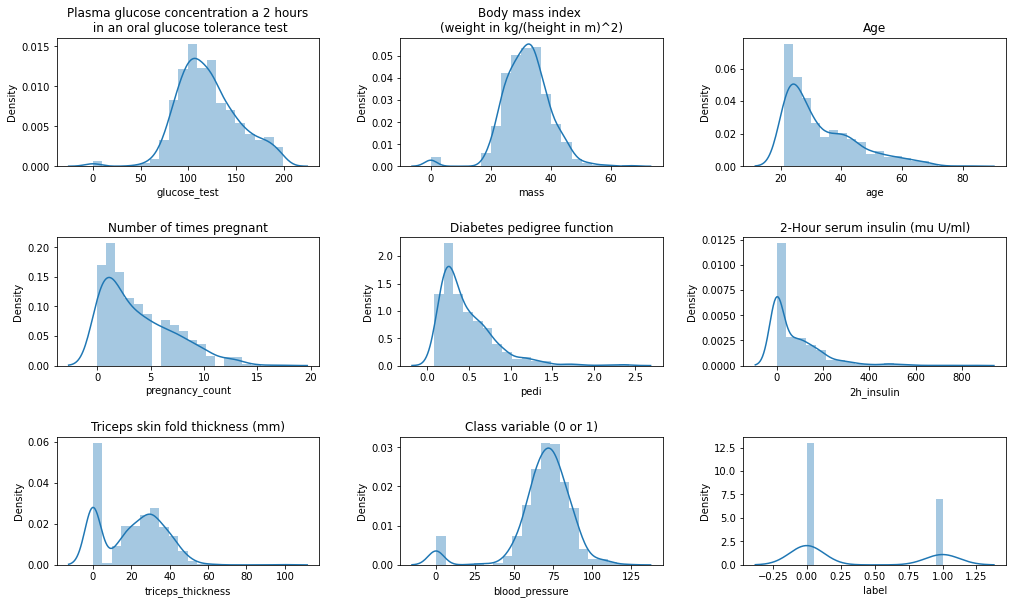
If we look closely we can say that body mass index and the plasma glucose is the most similar to normal distribution.
fig, ax = plt.subplots(3, 3, figsize=(20,15))
fig.tight_layout(pad=5)
sns.scatterplot(df.glucose_test,df.mass, ax=ax[0,0])
ax[0,0].legend(['glucose_test And mass'])
sns.scatterplot(df.mass, df.age, ax=ax[0,1])
ax[0, 1].legend(['mass And age'])
sns.scatterplot(df.age, df.pregnancy_count, ax=ax[0,2])
ax[0, 2].legend(['age And pregnancy_count'])
sns.scatterplot(df.pregnancy_count, df.pedi, ax=ax[1,0])
ax[1, 0].legend(['pregnancy_count And predi'])
sns.scatterplot(df.pedi, df['2h_insulin'], ax=ax[1, 1])
ax[1, 1].legend(['predi And 2h_insulin'])
sns.scatterplot(df['2h_insulin'], df.triceps_thickness, ax=ax[1, 2])
ax[1, 2].legend(['2h_insulin And triceps_thickness'])
sns.scatterplot(df.triceps_thickness, df.blood_pressure, ax=ax[2, 0])
ax[2, 0].legend(['triceps_thickness And blood_pressure'])
ax[2,1].set_axis_off()
ax[2,2].set_axis_off()
plt.show()
/home/amin/.local/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
/home/amin/.local/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
/home/amin/.local/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
/home/amin/.local/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
/home/amin/.local/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
/home/amin/.local/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
/home/amin/.local/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(
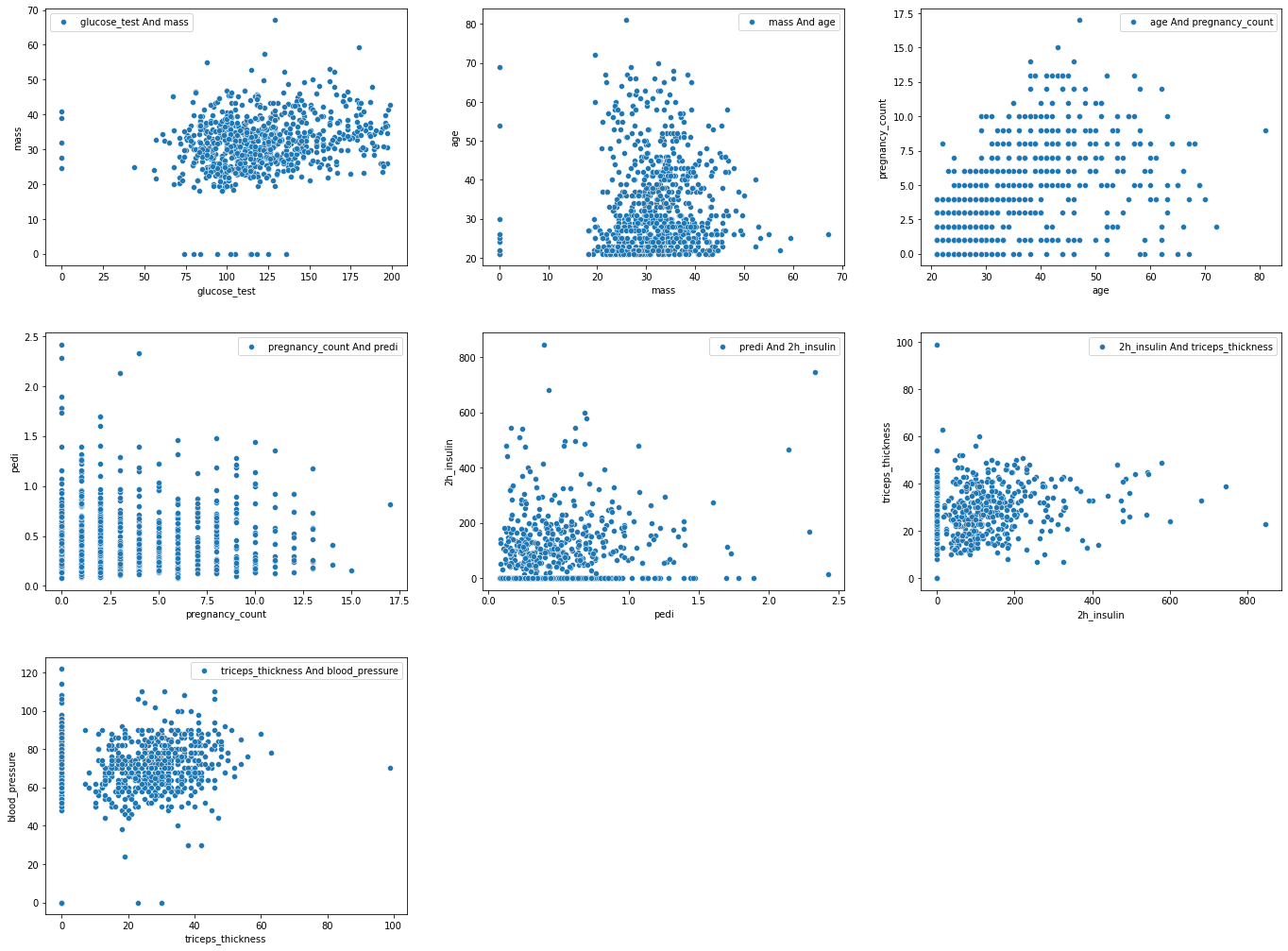
It is obvious that for (pregnancy_count, predi) and (age, pregnancy_count) we can say they have linear dependency.
Q3#
(a)#
create a normalize function using the equation below \begin{equation} x_{norm} = \frac{x- \mu_x}{\sigma_x} \end{equation} normalize the third attribute on pima dataset, then report the values for the first 5 entries on dataset.
## column names are used from ```pima-indians-diabetes.name``` file
cols = ['pregnancy_count', 'glucose_test', 'blood_pressure', 'triceps_thickness', '2h_insulin', 'mass', 'pedi', 'age', 'label']
df = pd.read_csv('hw1_data/pima/pima-indians-diabetes.data', index_col=False ,names=cols)
df.head()
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
def normalize(attrib):
"""
normalize an attribute using its mean and standard deviation
INPUTS:
--------
attrib: a pandas series, the original unormalized attribute
OUTPUT:
--------
normalized: the normalized attribute
"""
mu = attrib.mean()
sigma = attrib.std()
normalized = (attrib - mu) / sigma
return normalized
## third attribute is blood pressure
blood_pressure_norm = normalize(df.blood_pressure)
pd.DataFrame(blood_pressure_norm)
| blood_pressure | |
|---|---|
| 0 | 0.149543 |
| 1 | -0.160441 |
| 2 | -0.263769 |
| 3 | -0.160441 |
| 4 | -1.503707 |
| ... | ... |
| 763 | 0.356200 |
| 764 | 0.046215 |
| 765 | 0.149543 |
| 766 | -0.470426 |
| 767 | 0.046215 |
768 rows × 1 columns
## normalize first 5 attribute of the function
normalize(df[['pregnancy_count', 'glucose_test', 'blood_pressure', 'triceps_thickness', '2h_insulin']])
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | |
|---|---|---|---|---|---|
| 0 | 0.639530 | 0.847771 | 0.149543 | 0.906679 | -0.692439 |
| 1 | -0.844335 | -1.122665 | -0.160441 | 0.530556 | -0.692439 |
| 2 | 1.233077 | 1.942458 | -0.263769 | -1.287373 | -0.692439 |
| 3 | -0.844335 | -0.997558 | -0.160441 | 0.154433 | 0.123221 |
| 4 | -1.141108 | 0.503727 | -1.503707 | 0.906679 | 0.765337 |
| ... | ... | ... | ... | ... | ... |
| 763 | 1.826623 | -0.622237 | 0.356200 | 1.721613 | 0.869464 |
| 764 | -0.547562 | 0.034575 | 0.046215 | 0.405181 | -0.692439 |
| 765 | 0.342757 | 0.003299 | 0.149543 | 0.154433 | 0.279412 |
| 766 | -0.844335 | 0.159683 | -0.470426 | -1.287373 | -0.692439 |
| 767 | -0.844335 | -0.872451 | 0.046215 | 0.655930 | -0.692439 |
768 rows × 5 columns
new_df = df[['pregnancy_count', 'glucose_test', 'triceps_thickness', '2h_insulin']].copy()
new_df['normalized_blood_pressure'] = blood_pressure_norm
new_df.head()
| pregnancy_count | glucose_test | triceps_thickness | 2h_insulin | normalized_blood_pressure | |
|---|---|---|---|---|---|
| 0 | 6 | 148 | 35 | 0 | 0.149543 |
| 1 | 1 | 85 | 29 | 0 | -0.160441 |
| 2 | 8 | 183 | 0 | 0 | -0.263769 |
| 3 | 1 | 89 | 23 | 94 | -0.160441 |
| 4 | 0 | 137 | 35 | 168 | -1.503707 |
(b)#
def discretize_attribute(attribute ,bins=10, verbose=False):
"""
descritize a continues attribute and make the intervals the bins size
INPUTS:
--------
attribute: pandas series of **one column**
bins: the interval space for each class, default is 10
verbose: print the each stage output if True, default is False
OUTPUT:
--------
pandas dataframe of the discrite values of an attribute
"""
list = np.empty((1,len(attribute.columns)))
## each bin length
bin_len = attribute.max().max() / bins
# bin_len = int(bin_len)
if verbose: print(f'bin length: {bin_len}')
## counter to iterate the intervals
interval = 0
while interval < attribute.max().max():
## find the attributes within the interval and drop na (NAN values are thoes who are not in the interval)
conditioned_data = attribute.where((interval < attribute) & (attribute < interval + bin_len)).dropna().values
if verbose: print(conditioned_data)
## To discretize the attributes set all the values to the mean of interval we have
conditioned_data[True] = (interval + bin_len) / 2
list = np.append(list, conditioned_data, axis=0 )
## iterate the next intervals
interval += bin_len
return pd.DataFrame(list, columns=attribute.columns, dtype='float32')
attrib = df[['blood_pressure']]
df_discrete_blood_pressure = discretize_attribute(attrib, 10)
df_discrete_blood_pressure
| blood_pressure | |
|---|---|
| 0 | 0.000000 |
| 1 | 12.200000 |
| 2 | 18.299999 |
| 3 | 18.299999 |
| 4 | 24.400000 |
| ... | ... |
| 728 | 61.000000 |
| 729 | 61.000000 |
| 730 | 61.000000 |
| 731 | 61.000000 |
| 732 | 61.000000 |
733 rows × 1 columns
sns.histplot(df_discrete_blood_pressure)
plt.show()
df_discrete_pregnancy = discretize_attribute(df[['pregnancy_count']], 10)
df_discrete_glucose_test = discretize_attribute(df[['glucose_test']], 10)
df_discrete_triceps_thickness = discretize_attribute(df[['triceps_thickness']], 10)
df_discrete_2h_insulin = discretize_attribute(df[['2h_insulin']], 10)
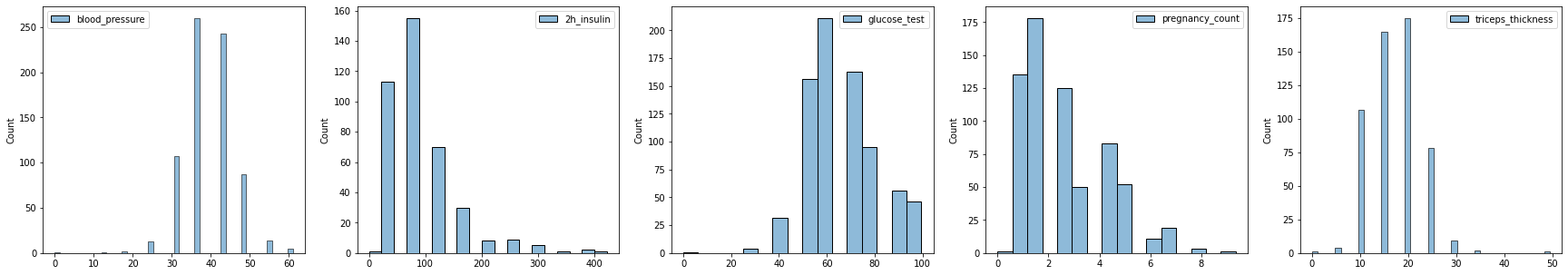
fig, axes = plt.subplots(1, 5, figsize=(30, 5))
sns.histplot(df_discrete_blood_pressure, ax=axes[0])
sns.histplot(df_discrete_2h_insulin, ax=axes[1])
sns.histplot(df_discrete_glucose_test, ax=axes[2])
sns.histplot(df_discrete_pregnancy, ax=axes[3])
sns.histplot(df_discrete_triceps_thickness, ax=axes[4])
plt.show()
Q4#
(a)#
## column names are used from ```pima-indians-diabetes.name``` file
cols = ['pregnancy_count', 'glucose_test', 'blood_pressure', 'triceps_thickness', '2h_insulin', 'mass', 'pedi', 'age', 'label']
df = pd.read_csv('hw1_data/pima/pima-indians-diabetes.data', index_col=False ,names=cols)
df.head()
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
df_class_0 = df[df.label == 0].copy()
df_class_1 = df[df.label == 1].copy()
df_class_0.describe()
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| count | 500.000000 | 500.0000 | 500.000000 | 500.000000 | 500.000000 | 500.000000 | 500.000000 | 500.000000 | 500.0 |
| mean | 3.298000 | 109.9800 | 68.184000 | 19.664000 | 68.792000 | 30.304200 | 0.429734 | 31.190000 | 0.0 |
| std | 3.017185 | 26.1412 | 18.063075 | 14.889947 | 98.865289 | 7.689855 | 0.299085 | 11.667655 | 0.0 |
| min | 0.000000 | 0.0000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.078000 | 21.000000 | 0.0 |
| 25% | 1.000000 | 93.0000 | 62.000000 | 0.000000 | 0.000000 | 25.400000 | 0.229750 | 23.000000 | 0.0 |
| 50% | 2.000000 | 107.0000 | 70.000000 | 21.000000 | 39.000000 | 30.050000 | 0.336000 | 27.000000 | 0.0 |
| 75% | 5.000000 | 125.0000 | 78.000000 | 31.000000 | 105.000000 | 35.300000 | 0.561750 | 37.000000 | 0.0 |
| max | 13.000000 | 197.0000 | 122.000000 | 60.000000 | 744.000000 | 57.300000 | 2.329000 | 81.000000 | 0.0 |
df_class_1.describe()
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| count | 268.000000 | 268.000000 | 268.000000 | 268.000000 | 268.000000 | 268.000000 | 268.000000 | 268.000000 | 268.0 |
| mean | 4.865672 | 141.257463 | 70.824627 | 22.164179 | 100.335821 | 35.142537 | 0.550500 | 37.067164 | 1.0 |
| std | 3.741239 | 31.939622 | 21.491812 | 17.679711 | 138.689125 | 7.262967 | 0.372354 | 10.968254 | 0.0 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.088000 | 21.000000 | 1.0 |
| 25% | 1.750000 | 119.000000 | 66.000000 | 0.000000 | 0.000000 | 30.800000 | 0.262500 | 28.000000 | 1.0 |
| 50% | 4.000000 | 140.000000 | 74.000000 | 27.000000 | 0.000000 | 34.250000 | 0.449000 | 36.000000 | 1.0 |
| 75% | 8.000000 | 167.000000 | 82.000000 | 36.000000 | 167.250000 | 38.775000 | 0.728000 | 44.000000 | 1.0 |
| max | 17.000000 | 199.000000 | 114.000000 | 99.000000 | 846.000000 | 67.100000 | 2.420000 | 70.000000 | 1.0 |
(b)#
## we would choose randomly an index with the probability of 66%
def divideset1(df, prob=0.66):
"""
divide the dataset into train and test with a probability
INPUTS:
--------
df: pandas dataframe, the dataset we want to split
prob: the probability to divide the dataset, default is 0.66
OUTPUTS:
---------
train: pandas dataframe, the portion of the dataset for train
test: pandas dataframe, the portion of the dataset for test
"""
## copy the dataframe to ensure there is no problem
dataset = df.copy()
trainset = pd.DataFrame(columns=dataset.columns)
testset = pd.DataFrame(columns=dataset.columns)
## iterate over dataset and select each row as train or test
for i in range(0, len(dataset)):
## get the row
row = dataset.iloc[0]
probability = np.random.random()
## if the probability range is between 0 and chosen prob
if probability - prob > 0:
trainset = trainset.append(row)
else:
testset = testset.append(row)
return trainset, testset
train_split, test_split = divideset1(df)
train_split
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
| 0 | 6.0 | 148.0 | 72.0 | 35.0 | 0.0 | 33.6 | 0.627 | 50.0 | 1.0 |
249 rows × 9 columns
## check the average training split size with the count of times
times = 20
## save all lengths
lengths = []
for i in range(times):
training_len = len(divideset1(df)[0])
print(f'Iteration {i} - training length: {training_len}')
lengths.append(training_len)
print('------------------------------------------')
print(f'Average Lengths: {np.average(lengths)}')
Iteration 0 - training length: 255
Iteration 1 - training length: 256
Iteration 2 - training length: 273
Iteration 3 - training length: 250
Iteration 4 - training length: 244
Iteration 5 - training length: 237
Iteration 6 - training length: 250
Iteration 7 - training length: 247
Iteration 8 - training length: 257
Iteration 9 - training length: 259
Iteration 10 - training length: 240
Iteration 11 - training length: 238
Iteration 12 - training length: 270
Iteration 13 - training length: 267
Iteration 14 - training length: 248
Iteration 15 - training length: 248
Iteration 16 - training length: 273
Iteration 17 - training length: 257
Iteration 18 - training length: 259
Iteration 19 - training length: 247
------------------------------------------
Average Lengths: 253.75
(c)#
def divideset2(df, fraction = 0.66):
"""
Divide the dataset into train and test with fixed size every run
INPUTS:
---------
df: pandas dataframe, the dataset that is going to be splitted
fraction: the value to divide the dataset, default is 0.66
OUTPUTS:
---------
train: pandas dataframe, the portion of the dataset for train
test: pandas dataframe, the portion of the dataset for test
"""
train = df.sample(frac=0.66).copy()
test = df.drop(train.index)
return train, test
train2, test2 = divideset2(df)
print(train2.head())
print('-----------------------'* 10)
print(test2.head())
pregnancy_count glucose_test blood_pressure triceps_thickness \
630 7 114 64 0
136 0 100 70 26
468 8 120 0 0
728 2 175 88 0
382 1 109 60 8
2h_insulin mass pedi age label
630 0 27.4 0.732 34 1
136 50 30.8 0.597 21 0
468 0 30.0 0.183 38 1
728 0 22.9 0.326 22 0
382 182 25.4 0.947 21 0
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
pregnancy_count glucose_test blood_pressure triceps_thickness \
0 6 148 72 35
2 8 183 64 0
3 1 89 66 23
6 3 78 50 32
11 10 168 74 0
2h_insulin mass pedi age label
0 0 33.6 0.627 50 1
2 0 23.3 0.672 32 1
3 94 28.1 0.167 21 0
6 88 31.0 0.248 26 1
11 0 38.0 0.537 34 1
Validation schemes#
(a) K-fold Cross Validation#
def kfold_crossvalidation(data, k, m):
"""
K-fold cross validation
Note: test data is equivalent the validation data in normal machine learning models (Because it is used to evaluate one model)
INPUTS:
--------
data: pandas dataframe containing feature vectors as rows
k: positive integer, the number of folds
m: target output
train_split: the fraction of data that is used to split for training, default is 0.7
OUTPUTS:
---------
training_data: multi-dimensional array of training data, each index contains the dataset for K-fold number
test_data: multi-dimensional array of test data, each ```index+1``` contains the dataset for each K-fold number
"""
## get the length of data to split it
dataframe_size = len(data)
## find the length of each split
# training_size = int(dataframe_size * train_split)
# test_size = dataframe_size - training_size
## empty arrays to save data into it
training_data = []
test_data = []
## find the split size
split = int(dataframe_size / k)
## split the data into k-fold and add the folds into the arrays
for i in range(k):
start_idx = int(i*split)
end_idx = int((i+1)*split)
test = data.iloc[start_idx:end_idx].copy()
## add the label column to corresponding index
test['label'] = m.iloc[start_idx: end_idx]
## choose other part of dataset as train
train = pd.concat([data, test, test]).drop_duplicates(keep=False)
train['label'] = m.iloc[train.index]
training_data.append(train)
test_data.append(test)
return training_data, test_data
K = 5
training_folds, test_folds = kfold_crossvalidation(df, K, df.label )
## check the K=1 test data fold
test_folds[0]
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 148 | 5 | 147 | 78 | 0 | 0 | 33.7 | 0.218 | 65 | 0 |
| 149 | 2 | 90 | 70 | 17 | 0 | 27.3 | 0.085 | 22 | 0 |
| 150 | 1 | 136 | 74 | 50 | 204 | 37.4 | 0.399 | 24 | 0 |
| 151 | 4 | 114 | 65 | 0 | 0 | 21.9 | 0.432 | 37 | 0 |
| 152 | 9 | 156 | 86 | 28 | 155 | 34.3 | 1.189 | 42 | 1 |
153 rows × 9 columns
## check K=1 fold training set
training_folds[0]
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 153 | 1 | 153 | 82 | 42 | 485 | 40.6 | 0.687 | 23 | 0 |
| 154 | 8 | 188 | 78 | 0 | 0 | 47.9 | 0.137 | 43 | 1 |
| 155 | 7 | 152 | 88 | 44 | 0 | 50.0 | 0.337 | 36 | 1 |
| 156 | 2 | 99 | 52 | 15 | 94 | 24.6 | 0.637 | 21 | 0 |
| 157 | 1 | 109 | 56 | 21 | 135 | 25.2 | 0.833 | 23 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 763 | 10 | 101 | 76 | 48 | 180 | 32.9 | 0.171 | 63 | 0 |
| 764 | 2 | 122 | 70 | 27 | 0 | 36.8 | 0.340 | 27 | 0 |
| 765 | 5 | 121 | 72 | 23 | 112 | 26.2 | 0.245 | 30 | 0 |
| 766 | 1 | 126 | 60 | 0 | 0 | 30.1 | 0.349 | 47 | 1 |
| 767 | 1 | 93 | 70 | 31 | 0 | 30.4 | 0.315 | 23 | 0 |
615 rows × 9 columns
## we can see that the test and train summation size matches the whole dataframe
for k in range(K):
## does it maches the whole dataset size? (condition variable)
condition = (len(df) == (len(training_folds[k]) + len(test_folds[k])))
print(f'Fold K={k+1}, the summation matches the whole set, {condition}')
Fold K=1, the summation matches the whole set, True
Fold K=2, the summation matches the whole set, True
Fold K=3, the summation matches the whole set, True
Fold K=4, the summation matches the whole set, True
Fold K=5, the summation matches the whole set, True
(b) Bootstraping#
def bootstrap1(data):
"""
Demonstrate one iteration of bootstraping method (it is a with replacement method)
INPUT:
-------
data: a pandas dataframe, containing our data
OUPUTS:
--------
train_data: pandas dataframe of a sample data
test_data: pandas dataframe of sample data, the data that are not included in train_data
"""
## find the length of our data (how many data rows we have)
data_length = len(data)
## the indexes to be chosen from original data
indexes = np.random.randint(data_length, size=data_length)
## create the training set
train_data = df.iloc[indexes].copy()
## choose the test set, (The data that is omited from training set)
test_data = pd.concat([data,train_data, train_data]).drop_duplicates(keep=False)
return train_data, test_data
bootstrap_train, bootstrap_test = bootstrap1(df)
## check the training set length with original data
len(bootstrap_train) == len(df)
True
print(len(bootstrap_train))
bootstrap_train.head()
768
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 455 | 14 | 175 | 62 | 30 | 0 | 33.6 | 0.212 | 38 | 1 |
| 67 | 2 | 109 | 92 | 0 | 0 | 42.7 | 0.845 | 54 | 0 |
| 692 | 2 | 121 | 70 | 32 | 95 | 39.1 | 0.886 | 23 | 0 |
| 191 | 9 | 123 | 70 | 44 | 94 | 33.1 | 0.374 | 40 | 0 |
| 711 | 5 | 126 | 78 | 27 | 22 | 29.6 | 0.439 | 40 | 0 |
print(len(bootstrap_test))
bootstrap_test.head()
277
| pregnancy_count | glucose_test | blood_pressure | triceps_thickness | 2h_insulin | mass | pedi | age | label | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 8 | 2 | 197 | 70 | 45 | 543 | 30.5 | 0.158 | 53 | 1 |
| 12 | 10 | 139 | 80 | 0 | 0 | 27.1 | 1.441 | 57 | 0 |
| 16 | 0 | 118 | 84 | 47 | 230 | 45.8 | 0.551 | 31 | 1 |
Q7#
(f)#
def poisson_distribution(X,lambda1):
"""
poisson distribution function
INPUT:
--------
X: integer or an array of integers, the input value
lambda1: float, hyperparameter to set
OUTPUT:
---------
probabiltiy: type is same as input, the probability distribution of X
"""
## calculate the value above the division and below separately
above_division = np.exp(- lambda1) * np.power(lambda1, X)
under_division = scipy.special.factorial(X)
return np.divide(above_division, under_division)
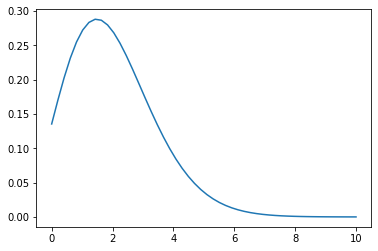
## parameter lambda = 2
X = np.linspace(0, 10, 50)
Y1 = poisson_distribution(X, lambda1=2)
plt.plot(X, Y1)
plt.show()
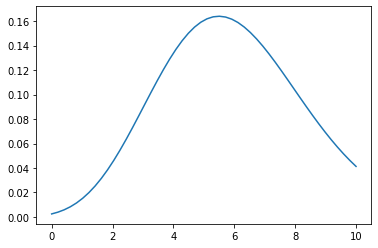
## parameter lambda = 6
X = np.linspace(0, 10, 50)
Y1 = poisson_distribution(X, lambda1=6)
plt.plot(X, Y1)
plt.show()
(g)#
The Maximum Likelihood estimation for poisson distribution is as below (Calculated in Q7 part c) \begin{equation} \lambda = \frac{1}{n} \sum_{i=0}^{n} x_i \end{equation}
## Read the data from poisson.txt file
X_poisson = np.fromfile('hw1_data/poisson.txt', dtype=float, sep='\n')
## Maximum Likelihood estimation
lambda1 = np.sum(X_poisson) / len(X_poisson)
print('Maximum Likelihood Estimated Parameter for poisson.txt: ', lambda1)
Maximum Likelihood Estimated Parameter for poisson.txt: 5.24
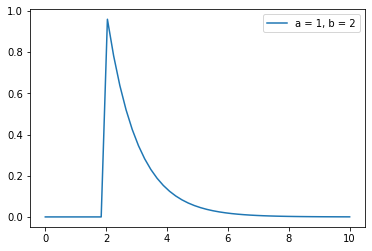
(h)#
X = np.linspace(0, 10, 50)
gamma_model = scipy.stats.gamma(1,2)
plt.plot(X, gamma_model.pdf(X))
plt.legend(['a = 1, b = 2'])
plt.show()
X = np.linspace(0, 10, 50)
gamma_model = scipy.stats.gamma(3,5)
plt.plot(X, gamma_model.pdf(X))
plt.legend(['a = 3, b = 5'])
plt.show()
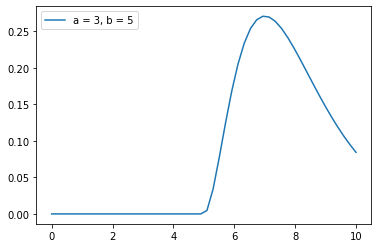
(i)#
Posterior density can be found as \begin{equation} Posterior = Likelihood \times Prior \end{equation} The prior of probability is \begin{equation} P(x) = \frac{e^{-\lambda} \lambda^x}{x!} \end{equation} \begin{equation} p(\lambda | a,b) = \frac{1}{b^a \Gamma(a)} \lambda^{a-1} e^{-\frac{\lambda}{b}} \end{equation} And assuming the poisson.txt file we found the proper value for lambda
def posterior_poisson(X, a,b, lambda1):
"""
posterior for poisson distribution
INPUTS:
-------
X: values to calculate the posterior
a,b: parameters for gamma distribution
OUTPUT:
--------
probability: floating number or an array
"""
## we divided the function in 3 parts then multiplied it
gamma_distro = scipy.stats.gamma(a)
values = gamma_distro.pdf(X)
p1 = (b ** a) * values
p1 = 1 / p1
p2 = lambda1 ** (a-1)
p3 = np.exp(-lambda1 / b)
probability = p1 * p2 * p3
print(len(probability))
return probability
Y = posterior_poisson(X_poisson, 3, 5, lambda1)
plt.figure(figsize=(8,6))
plt.plot(X_poisson,Y)
plt.legend(['Posterior distribution with a=1, b=2'])
plt.show()
25
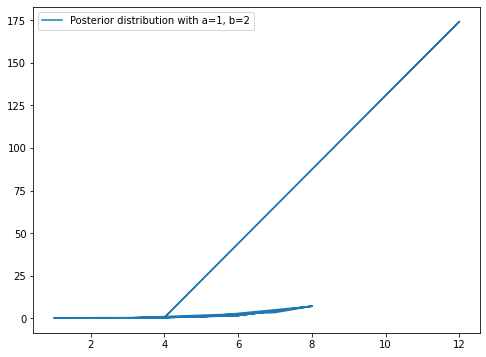
It can be seen from the plot that the posterior is not a function and it have multiple values of x for each y in range of 4 to 8.
Q8#
Part 1#
(a)#
## read the data from housing.txt
## there is one or more space for each data
columns = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD','TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
df_boston = pd.read_csv('hw1_data/housing/housing.txt', sep= ' +', index_col=False, names=columns)
/home/amin/.local/lib/python3.8/site-packages/pandas/util/_decorators.py:311: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support regex separators (separators > 1 char and different from '\s+' are interpreted as regex); you can avoid this warning by specifying engine='python'.
return func(*args, **kwargs)
df_boston.head()
| CRIM | ZN | INDUS | CHAS | NOX | RM | AGE | DIS | RAD | TAX | PTRATIO | B | LSTAT | MEDV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.00632 | 18.0 | 2.31 | 0 | 0.538 | 6.575 | 65.2 | 4.0900 | 1 | 296.0 | 15.3 | 396.90 | 4.98 | 24.0 |
| 1 | 0.02731 | 0.0 | 7.07 | 0 | 0.469 | 6.421 | 78.9 | 4.9671 | 2 | 242.0 | 17.8 | 396.90 | 9.14 | 21.6 |
| 2 | 0.02729 | 0.0 | 7.07 | 0 | 0.469 | 7.185 | 61.1 | 4.9671 | 2 | 242.0 | 17.8 | 392.83 | 4.03 | 34.7 |
| 3 | 0.03237 | 0.0 | 2.18 | 0 | 0.458 | 6.998 | 45.8 | 6.0622 | 3 | 222.0 | 18.7 | 394.63 | 2.94 | 33.4 |
| 4 | 0.06905 | 0.0 | 2.18 | 0 | 0.458 | 7.147 | 54.2 | 6.0622 | 3 | 222.0 | 18.7 | 396.90 | 5.33 | 36.2 |
df_boston.dtypes
CRIM float64
ZN float64
INDUS float64
CHAS int64
NOX float64
RM float64
AGE float64
DIS float64
RAD int64
TAX float64
PTRATIO float64
B float64
LSTAT float64
MEDV float64
dtype: object
As we can see there is no attribute type as object, so we will look at the dataset closely.
df_boston.CHAS.value_counts()
0 471
1 35
Name: CHAS, dtype: int64
It is clear here that CHAS is a binary feature.
df_boston.ZN.value_counts()
0.0 372
20.0 21
80.0 15
22.0 10
12.5 10
25.0 10
40.0 7
45.0 6
30.0 6
90.0 5
95.0 4
60.0 4
21.0 4
33.0 4
55.0 3
70.0 3
34.0 3
52.5 3
35.0 3
28.0 3
75.0 3
82.5 2
85.0 2
17.5 1
100.0 1
18.0 1
Name: ZN, dtype: int64
So after having a look at dataset and two of the features we saw that just one of the columns have binary value, and it is the CHAS.
(b)#
correlation = df_boston[columns[:-1]].corrwith(df_boston.MEDV).sort_values(ascending=True)
pd.DataFrame(correlation, columns=['correlation with MEDV'])
| correlation with MEDV | |
|---|---|
| LSTAT | -0.737663 |
| PTRATIO | -0.507787 |
| INDUS | -0.483725 |
| TAX | -0.468536 |
| NOX | -0.427321 |
| CRIM | -0.388305 |
| RAD | -0.381626 |
| AGE | -0.376955 |
| CHAS | 0.175260 |
| DIS | 0.249929 |
| B | 0.333461 |
| ZN | 0.360445 |
| RM | 0.695360 |
It is clear from above that:
Highest Positive Correlation: RM
Highest Negative Correlation: LSTAT
(c)#
fig, axes = plt.subplots(3, 5, figsize=(25, 10))
fig.tight_layout(pad=5)
sns.scatterplot(x=df_boston.LSTAT, y=df_boston.MEDV, ax=axes[0,0])
sns.scatterplot(x=df_boston.PTRATIO, y=df_boston.MEDV, ax=axes[0,1])
sns.scatterplot(x=df_boston.INDUS, y=df_boston.MEDV, ax=axes[0,2])
sns.scatterplot(x=df_boston.TAX, y=df_boston.MEDV, ax=axes[0,3])
sns.scatterplot(x=df_boston.NOX, y=df_boston.MEDV, ax=axes[0,4])
sns.scatterplot(x=df_boston.CRIM, y=df_boston.MEDV, ax=axes[1,0])
sns.scatterplot(x=df_boston.AGE, y=df_boston.MEDV, ax=axes[1,1])
sns.scatterplot(x=df_boston.CHAS, y=df_boston.MEDV, ax=axes[1,2])
sns.scatterplot(x=df_boston.DIS, y=df_boston.MEDV, ax=axes[1,3])
sns.scatterplot(x=df_boston.B, y=df_boston.MEDV, ax=axes[1,4])
sns.scatterplot(x=df_boston.ZN, y=df_boston.MEDV, ax=axes[2,0])
sns.scatterplot(x=df_boston.RM, y=df_boston.MEDV, ax=axes[2,1])
axes[2,2].set_axis_off()
axes[2,3].set_axis_off()
axes[2,4].set_axis_off()
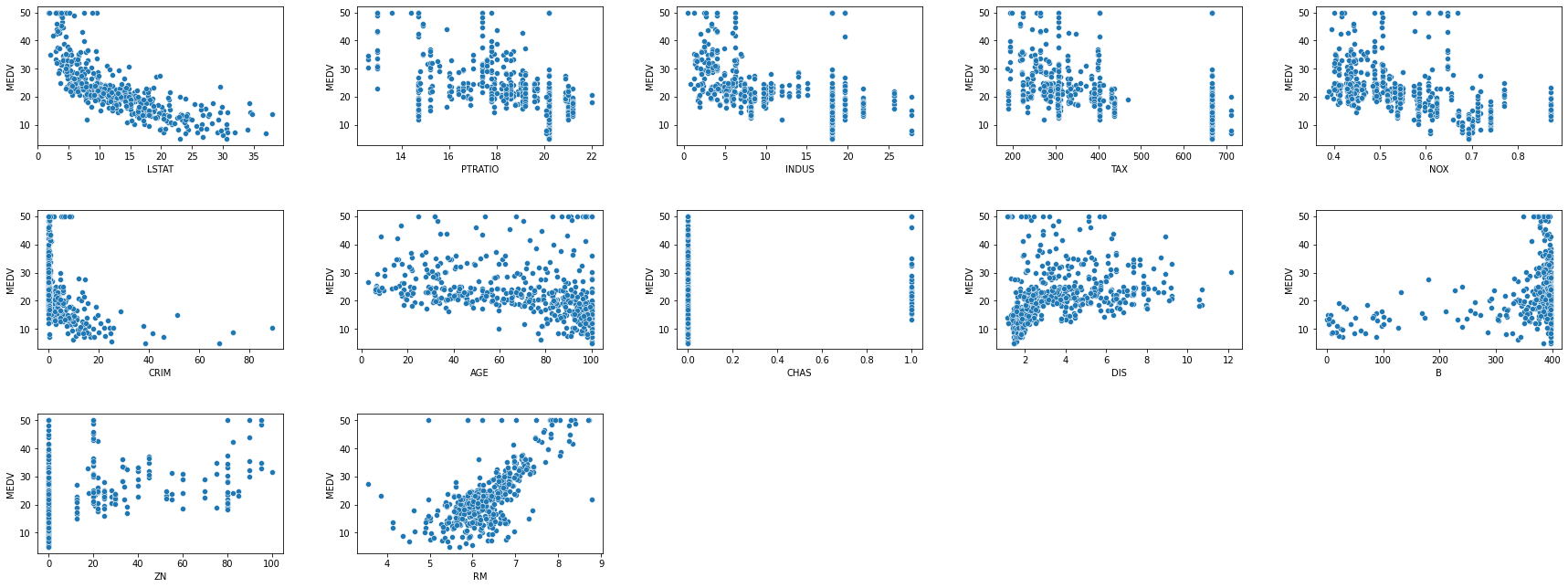
The most correlated features are having the most linear scatter plot. As we can see from the scatter plots, RM and LSTAT are in the most linear form.
(d)#
df_boston.corr()
| CRIM | ZN | INDUS | CHAS | NOX | RM | AGE | DIS | RAD | TAX | PTRATIO | B | LSTAT | MEDV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRIM | 1.000000 | -0.200469 | 0.406583 | -0.055892 | 0.420972 | -0.219247 | 0.352734 | -0.379670 | 0.625505 | 0.582764 | 0.289946 | -0.385064 | 0.455621 | -0.388305 |
| ZN | -0.200469 | 1.000000 | -0.533828 | -0.042697 | -0.516604 | 0.311991 | -0.569537 | 0.664408 | -0.311948 | -0.314563 | -0.391679 | 0.175520 | -0.412995 | 0.360445 |
| INDUS | 0.406583 | -0.533828 | 1.000000 | 0.062938 | 0.763651 | -0.391676 | 0.644779 | -0.708027 | 0.595129 | 0.720760 | 0.383248 | -0.356977 | 0.603800 | -0.483725 |
| CHAS | -0.055892 | -0.042697 | 0.062938 | 1.000000 | 0.091203 | 0.091251 | 0.086518 | -0.099176 | -0.007368 | -0.035587 | -0.121515 | 0.048788 | -0.053929 | 0.175260 |
| NOX | 0.420972 | -0.516604 | 0.763651 | 0.091203 | 1.000000 | -0.302188 | 0.731470 | -0.769230 | 0.611441 | 0.668023 | 0.188933 | -0.380051 | 0.590879 | -0.427321 |
| RM | -0.219247 | 0.311991 | -0.391676 | 0.091251 | -0.302188 | 1.000000 | -0.240265 | 0.205246 | -0.209847 | -0.292048 | -0.355501 | 0.128069 | -0.613808 | 0.695360 |
| AGE | 0.352734 | -0.569537 | 0.644779 | 0.086518 | 0.731470 | -0.240265 | 1.000000 | -0.747881 | 0.456022 | 0.506456 | 0.261515 | -0.273534 | 0.602339 | -0.376955 |
| DIS | -0.379670 | 0.664408 | -0.708027 | -0.099176 | -0.769230 | 0.205246 | -0.747881 | 1.000000 | -0.494588 | -0.534432 | -0.232471 | 0.291512 | -0.496996 | 0.249929 |
| RAD | 0.625505 | -0.311948 | 0.595129 | -0.007368 | 0.611441 | -0.209847 | 0.456022 | -0.494588 | 1.000000 | 0.910228 | 0.464741 | -0.444413 | 0.488676 | -0.381626 |
| TAX | 0.582764 | -0.314563 | 0.720760 | -0.035587 | 0.668023 | -0.292048 | 0.506456 | -0.534432 | 0.910228 | 1.000000 | 0.460853 | -0.441808 | 0.543993 | -0.468536 |
| PTRATIO | 0.289946 | -0.391679 | 0.383248 | -0.121515 | 0.188933 | -0.355501 | 0.261515 | -0.232471 | 0.464741 | 0.460853 | 1.000000 | -0.177383 | 0.374044 | -0.507787 |
| B | -0.385064 | 0.175520 | -0.356977 | 0.048788 | -0.380051 | 0.128069 | -0.273534 | 0.291512 | -0.444413 | -0.441808 | -0.177383 | 1.000000 | -0.366087 | 0.333461 |
| LSTAT | 0.455621 | -0.412995 | 0.603800 | -0.053929 | 0.590879 | -0.613808 | 0.602339 | -0.496996 | 0.488676 | 0.543993 | 0.374044 | -0.366087 | 1.000000 | -0.737663 |
| MEDV | -0.388305 | 0.360445 | -0.483725 | 0.175260 | -0.427321 | 0.695360 | -0.376955 | 0.249929 | -0.381626 | -0.468536 | -0.507787 | 0.333461 | -0.737663 | 1.000000 |
The most correlated ones are TAX and RAD with the correlation value of 0.910228.
Part 2#
def df_housing_read(test = False):
columns = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD','TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
if test:
df = pd.read_csv('hw1_data/housing/housing_test.txt', sep=' +', names=columns, index_col=False, engine='python')
else:
df = pd.read_csv('hw1_data/housing/housing_train.txt', sep=' +', names=columns, index_col=False, engine='python')
return df
(a)#
df_train = df_housing_read()
df_train.head()
| CRIM | ZN | INDUS | CHAS | NOX | RM | AGE | DIS | RAD | TAX | PTRATIO | B | LSTAT | MEDV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.00632 | 18.0 | 2.31 | 0 | 0.538 | 6.575 | 65.2 | 4.0900 | 1 | 296.0 | 15.3 | 396.90 | 4.98 | 24.0 |
| 1 | 0.02731 | 0.0 | 7.07 | 0 | 0.469 | 6.421 | 78.9 | 4.9671 | 2 | 242.0 | 17.8 | 396.90 | 9.14 | 21.6 |
| 2 | 0.02729 | 0.0 | 7.07 | 0 | 0.469 | 7.185 | 61.1 | 4.9671 | 2 | 242.0 | 17.8 | 392.83 | 4.03 | 34.7 |
| 3 | 0.03237 | 0.0 | 2.18 | 0 | 0.458 | 6.998 | 45.8 | 6.0622 | 3 | 222.0 | 18.7 | 394.63 | 2.94 | 33.4 |
| 4 | 0.06905 | 0.0 | 2.18 | 0 | 0.458 | 7.147 | 54.2 | 6.0622 | 3 | 222.0 | 18.7 | 396.90 | 5.33 | 36.2 |
Target output is MEDV, and we can split it from the train dataset.
## Get feature vectors and target output
Y = df_train.MEDV.copy()
X = df_train.drop(['MEDV'], axis=1)
Linear regression weights can be found as \begin{equation} w = A^{-1} b \end{equation} where b and A can be found as \begin{equation} A = \sum_{i=0}^{n} x_i x_i^{T} \end{equation} \begin{equation} b = \sum_{i=0}^{n} y_i x_i \end{equation}
def LR_solve(X, Y):
"""
Linear regression solve function
Using the old equation w = invers(A) * b
Parameters:
--------
X : matrix_like
The feature vectors matrix
Y : array_like
The vector of each feature vector labels (target outputs)
Returns:
--------
w : array_like
The vector of weights fitted on `X` features
"""
A = X.dot(X.T)
## create b and preprocess it
b = Y.multiply(X)
b = np.sum(b, axis=1)
w = np.linalg.inv(A).dot(b)
return w
## giving the transpose of X, because it's as row in our dataset
w = LR_solve(X.T, Y)
print(f'Weights: {w}', f'\nShape:{w.shape}')
Weights: [-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
Shape:(13,)
(b)#
def LR_predict(X_test, w):
"""
Predict the Linear Regression using fixed input weights
Paramaters:
-----------
X_test : matrix_like
test data, an array of feature vectors
w : array_like
array of weights, shape must meet `(n, 1)`, column vector
Returns:
--------
Y_pred : array_like
the prediction of test data, using weights
"""
Y_pred = X_test @ w
return Y_pred
columns = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD','TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
df_test = pd.read_csv('hw1_data/housing/housing_test.txt', sep=' +', names=columns, index_col=False, engine='python')
df_test.head()
| CRIM | ZN | INDUS | CHAS | NOX | RM | AGE | DIS | RAD | TAX | PTRATIO | B | LSTAT | MEDV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.84054 | 0.0 | 8.14 | 0 | 0.538 | 5.599 | 85.7 | 4.4546 | 4 | 307.0 | 21.0 | 303.42 | 16.51 | 13.9 |
| 1 | 0.67191 | 0.0 | 8.14 | 0 | 0.538 | 5.813 | 90.3 | 4.6820 | 4 | 307.0 | 21.0 | 376.88 | 14.81 | 16.6 |
| 2 | 0.95577 | 0.0 | 8.14 | 0 | 0.538 | 6.047 | 88.8 | 4.4534 | 4 | 307.0 | 21.0 | 306.38 | 17.28 | 14.8 |
| 3 | 0.77299 | 0.0 | 8.14 | 0 | 0.538 | 6.495 | 94.4 | 4.4547 | 4 | 307.0 | 21.0 | 387.94 | 12.80 | 18.4 |
| 4 | 1.00245 | 0.0 | 8.14 | 0 | 0.538 | 6.674 | 87.3 | 4.2390 | 4 | 307.0 | 21.0 | 380.23 | 11.98 | 21.0 |
X_test = df_test.drop(columns=['MEDV'])
Y_test_actual = df_test.MEDV
Y_test_pred = LR_predict(X_test, w)
Y_test_pred[:5]
0 13.411778
1 16.500643
2 15.674173
3 21.839123
4 23.367941
dtype: float64
(c, d)#
## the main3_2.py program
!python3 scripts/main3_2.py
Linear Regression on housing dataset program!
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
Training:
Mean Squared Error: 24.475882784643673
Testing:
Mean Squared Error: 24.29223817565946
----------------------------------------------------------------------
Weights of the training
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
Part 3#
(a) Online Linear Regression#
The equation for online linear regression is \begin{equation} W_i = W_{i-1} + \alpha_i (y_i - f(x_i, W_{i-1})) x_i \end{equation} Where \(\alpha\) is the learning rate, \(W_i\) is the weights rate fot the \(i\)-th iteration and \(x_i\) is the \(i\)-th feature vector.
df_train = df_housing_read()
def normalize(df):
"""
normalize a pandas dataframe
Parameters:
------------
df : pandas dataframe
the dataset that is going to be normalized
Returns:
----------
df : pandas dataframe
the normalized dataframe
"""
df_normalized = df.copy()
cols = df_normalized.columns
for col in cols:
df_normalized[col] = (df_normalized[col] - df_normalized[col].mean() ) / df_normalized[col].std()
return df_normalized
df_train_normal = normalize(df_train)
df_train_normal.head()
| CRIM | ZN | INDUS | CHAS | NOX | RM | AGE | DIS | RAD | TAX | PTRATIO | B | LSTAT | MEDV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.406074 | 0.271666 | -1.269381 | -0.267612 | -0.128607 | 0.384651 | -0.095048 | 0.140689 | -0.969579 | -0.612355 | -1.449420 | 0.409077 | -1.046568 | 0.119273 |
| 1 | -0.403616 | -0.486839 | -0.554501 | -0.267612 | -0.726487 | 0.168444 | 0.397814 | 0.567315 | -0.854463 | -0.934791 | -0.274757 | 0.409077 | -0.459782 | -0.133425 |
| 2 | -0.403618 | -0.486839 | -0.554501 | -0.267612 | -0.726487 | 1.241055 | -0.242547 | 0.567315 | -0.854463 | -0.934791 | -0.274757 | 0.360950 | -1.180570 | 1.245885 |
| 3 | -0.403023 | -0.486839 | -1.288905 | -0.267612 | -0.821802 | 0.978518 | -0.792968 | 1.099976 | -0.739347 | -1.054212 | 0.148121 | 0.382235 | -1.334319 | 1.109007 |
| 4 | -0.398727 | -0.486839 | -1.288905 | -0.267612 | -0.821802 | 1.187706 | -0.490776 | 1.099976 | -0.739347 | -1.054212 | 0.148121 | 0.409077 | -0.997199 | 1.403821 |
def LR_Incremental_solve(X, Y, W, iter = 1000):
"""
Incremental Learning for Linear Regression
The method used is online gradient descent
the learning rate is 2/t, and t stands for iteration number
Parameters:
------------
X : matrix_like
the features vectors for training
Y : array_like
the target output for each feature vectors represented in `X`
W : array_like
the initial weights for online gradient descent
iter : integer
the number of iterations to learn
Returns:
---------
W : array_like
the learned weights for linear regression
"""
for i in range(iter):
## data index is different from the index
## so we calculate it everytime
data_index = i % len(Y)
## the update term that is added to old weight
update_term = Y[data_index] - Function(X.iloc[data_index], W)
## the learning rate
learning_rate = (2/(i+1))
update_term = np.multiply(learning_rate * update_term, X.iloc[data_index])
## update the weights
W = np.add(W, update_term)
return W
def Function(X, W):
"""
The function for calculating the predicted output for `X`
Parameters:
-----------
X : array_like
the features vector
W : array_like
the learned weights
Returns:
--------
Y_pred : float
The predicted value for the input weights and the feature vector
"""
Y_pred = np.dot(W.T, X)
return Y_pred
## initialize the data
X_train = df_train_normal.drop(columns=['MEDV'])
Y_train = df_train_normal.MEDV
W = np.zeros(13)
weights = LR_Incremental_solve(X = X_train,Y= Y_train,W= W)
weights
CRIM -0.020193
ZN 0.620314
INDUS 0.289735
CHAS -0.076357
NOX 3.653932
RM 0.948070
AGE -1.926829
DIS 1.026664
RAD -2.659829
TAX 0.703401
PTRATIO 1.108539
B 1.023090
LSTAT -0.511682
dtype: float64
(b)#
!python3 scripts/main3_3.py
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 24.475882784643673
Testing using the last trained weights:
Test
Mean Squared Error: 24.29223817565946
----------------------------------------------------------------------
Weights of the training
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 6.794950630755434
Test
Mean Squared Error: 269174.89239789307
----------------------------------------------------------------------
Final wights of Incremental Learning
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
(c)#
Using unnormalized dataset, will overflow and the program would stop.
## we're using unnormalized dataset
X = df_train.drop(columns=['MEDV'])
Y = df_train.MEDV
W = np.zeros(13)
weights = LR_Incremental_solve(X = X,Y= Y,W= W)
weights
CRIM NaN
ZN NaN
INDUS NaN
CHAS NaN
NOX NaN
RM NaN
AGE NaN
DIS NaN
RAD NaN
TAX NaN
PTRATIO NaN
B NaN
LSTAT NaN
dtype: float64
Getting NaN shows that overflow has happened.
(d)#
!python3 scripts/main3_3.py
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 24.475882784643673
Testing using the last trained weights:
Test
Mean Squared Error: 24.29223817565946
----------------------------------------------------------------------
Weights of the training
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 6.794950630755434
Test
Mean Squared Error: 269174.89239789307
----------------------------------------------------------------------
Final wights of Incremental Learning
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
(d.2)#
## using the learning rate 2 / sqrt(t)
!python3 scripts/main3_3.py learning_rate=1
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 24.475882784643673
Testing using the last trained weights:
Test
Mean Squared Error: 24.29223817565946
----------------------------------------------------------------------
Weights of the training
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 1.5252088672442436e+54
Test
Mean Squared Error: 4.577866801177761e+56
----------------------------------------------------------------------
Final wights of Incremental Learning
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
## using static learning rate 0.5
!python3 scripts/main3_3.py learning_rate=0.5
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 24.475882784643673
Testing using the last trained weights:
Test
Mean Squared Error: 24.29223817565946
----------------------------------------------------------------------
Weights of the training
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 0.0
Test
Mean Squared Error: 0.0
----------------------------------------------------------------------
Final wights of Incremental Learning
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
## using static learning rate 0.5
!python3 scripts/main3_3.py learning_rate=0.01
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 24.475882784643673
Testing using the last trained weights:
Test
Mean Squared Error: 24.29223817565946
----------------------------------------------------------------------
Weights of the training
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 0.29961777287140706
Test
Mean Squared Error: 1568.3310594686245
----------------------------------------------------------------------
Final wights of Incremental Learning
[-9.79342380e-02 4.89586765e-02 -2.53928478e-02 3.45087927e+00
-3.55458931e-01 5.81653272e+00 -3.31447963e-03 -1.02050134e+00
2.26563208e-01 -1.22458785e-02 -3.88029879e-01 1.70214971e-02
-4.85012955e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
As we saw, the losses for both training set and test set online Linear Regression (LR) with learning rate 0.5 is zero. We can conclude from this, that the online LR with learning rate 0.5 can be the most suitable model for our problem.
Part 4#
(a)#
def extendx(X):
"""
extend the X dataset and return the linear and two degrees dataset
Parameters:
-----------
X : matrix_like
our dataset
Returns:
--------
extended_X : matrix_like
the dataset containing both X and X^2 data
Note: extended_X length is double the X
"""
df = X.copy()
## the power 2
df_2 = df.multiply(df)
df = pd.concat([ df ,df_2], ignore_index= True)
return df
df = df_housing_read()
extendx(df)
| CRIM | ZN | INDUS | CHAS | NOX | RM | AGE | DIS | RAD | TAX | PTRATIO | B | LSTAT | MEDV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.006320 | 18.0 | 2.3100 | 0 | 0.538000 | 6.575000 | 65.20 | 4.090000 | 1 | 296.0 | 15.3 | 396.9000 | 4.9800 | 24.00 |
| 1 | 0.027310 | 0.0 | 7.0700 | 0 | 0.469000 | 6.421000 | 78.90 | 4.967100 | 2 | 242.0 | 17.8 | 396.9000 | 9.1400 | 21.60 |
| 2 | 0.027290 | 0.0 | 7.0700 | 0 | 0.469000 | 7.185000 | 61.10 | 4.967100 | 2 | 242.0 | 17.8 | 392.8300 | 4.0300 | 34.70 |
| 3 | 0.032370 | 0.0 | 2.1800 | 0 | 0.458000 | 6.998000 | 45.80 | 6.062200 | 3 | 222.0 | 18.7 | 394.6300 | 2.9400 | 33.40 |
| 4 | 0.069050 | 0.0 | 2.1800 | 0 | 0.458000 | 7.147000 | 54.20 | 6.062200 | 3 | 222.0 | 18.7 | 396.9000 | 5.3300 | 36.20 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 861 | 0.003923 | 0.0 | 142.3249 | 0 | 0.328329 | 43.467649 | 4774.81 | 6.143458 | 1 | 74529.0 | 441.0 | 153656.1601 | 93.5089 | 501.76 |
| 862 | 0.002049 | 0.0 | 142.3249 | 0 | 0.328329 | 37.454400 | 5882.89 | 5.232656 | 1 | 74529.0 | 441.0 | 157529.6100 | 82.4464 | 424.36 |
| 863 | 0.003692 | 0.0 | 142.3249 | 0 | 0.328329 | 48.664576 | 8281.00 | 4.698056 | 1 | 74529.0 | 441.0 | 157529.6100 | 31.8096 | 571.21 |
| 864 | 0.012010 | 0.0 | 142.3249 | 0 | 0.328329 | 46.158436 | 7974.49 | 5.706843 | 1 | 74529.0 | 441.0 | 154802.9025 | 41.9904 | 484.00 |
| 865 | 0.002248 | 0.0 | 142.3249 | 0 | 0.328329 | 36.360900 | 6528.64 | 6.275025 | 1 | 74529.0 | 441.0 | 157529.6100 | 62.0944 | 141.61 |
866 rows × 14 columns
(b)#
For binary attributes, the transformation would act as \(and\) logical operator. Binary attributes in the part of dataset with Linear form won’t change, but in the polynomial the binaries will be logically \(and\).
(c)#
!python3 scripts/main3_4.py
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 55343.71451079023
Testing using the last trained weights:
Test
Mean Squared Error: 35394.311865625714
----------------------------------------------------------------------
Weights of the training
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 0.7264756830925618
Test
Mean Squared Error: 35097170.23443318
----------------------------------------------------------------------
Final wights of Incremental Learning
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
!python3 scripts/main3_4.py learning_rate=1
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 55343.71451079023
Testing using the last trained weights:
Test
Mean Squared Error: 35394.311865625714
----------------------------------------------------------------------
Weights of the training
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 2.9753029489387566e+38
Test
Mean Squared Error: 1.1151099259756206e+46
----------------------------------------------------------------------
Final wights of Incremental Learning
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
!python3 scripts/main3_4.py learning_rate=0.5
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 55343.71451079023
Testing using the last trained weights:
Test
Mean Squared Error: 35394.311865625714
----------------------------------------------------------------------
Weights of the training
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 0.0
Test
Mean Squared Error: 0.0
----------------------------------------------------------------------
Final wights of Incremental Learning
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
!python3 scripts/main3_4.py learning_rate=0.01
Linear Regression on housing dataset program!
----------------------------------------------------------------------
----------------------------------------------------------------------
Loading Training and Test sets
datasets loaded!
Train set head:
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.00632 18.0 2.31 0 0.538 ... 1 296.0 15.3 396.90 4.98
1 0.02731 0.0 7.07 0 0.469 ... 2 242.0 17.8 396.90 9.14
2 0.02729 0.0 7.07 0 0.469 ... 2 242.0 17.8 392.83 4.03
3 0.03237 0.0 2.18 0 0.458 ... 3 222.0 18.7 394.63 2.94
4 0.06905 0.0 2.18 0 0.458 ... 3 222.0 18.7 396.90 5.33
[5 rows x 13 columns]
Test set head :
CRIM ZN INDUS CHAS NOX ... RAD TAX PTRATIO B LSTAT
0 0.84054 0.0 8.14 0 0.538 ... 4 307.0 21.0 303.42 16.51
1 0.67191 0.0 8.14 0 0.538 ... 4 307.0 21.0 376.88 14.81
2 0.95577 0.0 8.14 0 0.538 ... 4 307.0 21.0 306.38 17.28
3 0.77299 0.0 8.14 0 0.538 ... 4 307.0 21.0 387.94 12.80
4 1.00245 0.0 8.14 0 0.538 ... 4 307.0 21.0 380.23 11.98
[5 rows x 13 columns]
----------------------------------------------------------------------
----------------------------------------------------------------------
Training:
Mean Squared Error: 55343.71451079023
Testing using the last trained weights:
Test
Mean Squared Error: 35394.311865625714
----------------------------------------------------------------------
Weights of the training
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
----------------------------------------------------------------------
Incremental Learning
Training
Mean Squared Error: 0.31370632734922127
Test
Mean Squared Error: 89882561.46377167
----------------------------------------------------------------------
Final wights of Incremental Learning
[-4.68476601e-02 4.08283559e-02 8.30964818e-02 1.09347853e+02
-2.39244676e+02 3.05351274e+01 2.35384427e-03 -5.57037854e+00
8.67934356e-01 -1.28788975e-03 -1.50542358e+00 1.40912747e-03
-3.64281060e-01]
----------------------------------------------------------------------
Partial Wights
Plotting Training and Test Loss
Figure(1500x500)
(d)#
Comparing the plotted losses shows that there is no big differences between both parts c and d, but there are some minor differences comparing between them. For example using the \(\frac{2}{t}\) learning rate shows that the extended version of the dataset achieves the best results later than the linear version. But in contrast we can say that the we can achieve better results with linear version in more early steps.
